The use of Python for data science and analytics is growing in popularity and one reason for this is the excellent supporting libraries (NumPy, SciPy, pandas, Statsmodels (ref), Scikit-Learn, and Matplotlib, to name the most common ones). One obstacle to adoption can be lack of documentation: e.g. Statsmodels documentation is sparse and assumes a fair level of statistical knowledge to make use of it. This article shows how one feature of Statsmodels, namely Generalized Linear Models (GLM), can be used to build useful models for understanding count data.
{% include mathjax.html %}
As the name implies, Statsmodels provides statistical modelling capabilities in Python with a particular strength in econometric analysis. It builds on the capabilities of NumPy/SciPy for numerical array handling and scientific functions. It also integrates with pandas for input of data tables. On that foundation it delivers a range of statistical models with a straightforward but flexible API. Statsmodels has modules for exploration, testing and estimation across a broad range of models, but for this article we are interested in regression.
Prior to this project I was aware of Generalized Linear Models (ref), but had not had a suitable opportunity to try them. In the process I learned how straightforward this is using Statsmodels’ GLM module.
The Problem At Hand
As part of a client engagement we were examining beverage sales for a hotel in inner-suburban Melbourne. The larger goal was to explore the influence of various factors on patrons’ beverage consumption, including music, weather, time of day/week and local events. The investigation was not part of a planned experiment, rather it was an exploratory analysis of available historical data to see if there might be any discernible effect of these factors.
The rate of sales in a public bar can vary enormously between quiet and peak times. Evenings and week-ends are busier and local public events that attract a crowd can move patrons into and out of the venue at certain times. For this article we are exploring a subset of the data covering the quieter days, Monday – Thursday, and just the hour 6pm – 7pm. There are 126 records and their distribution is shown in Figure 1 below. We immediately note there is a small number of large values which may not be easy to accommodate in a model.
Figure 1. Beverage sales

We start by importing the dataset into a pandas DataFrame and after some exploration decide on the following columns. (pandas makes importing easy – see the documentation for examples.)
Table 1: DataFrame schema
| Column | Description |
|---|---|
| bev_quant | Number of drinks sold for the hour |
| bpm | Median music temp (beats per min) |
| hour | Timestamp with date at the end of the hour |
| is_happy_hr | Flag indicating a Happy Hour with discount drinks |
| is_mon_pubhol | Flag indicating day is a Monday public holiday |
| is_pre_xmas | Flag indicating day lies in the 2-week, pre-Christmas period |
| is_pubhol | Flag indicating a public holiday |
| is_pubhol_eve | Flag indicating the eve of a public holiday |
| n_events | Number of local events before/after hour |
| precip | Precipitation (mm) |
| relhum | Relative humidity (%) |
| temp | Temperature (Celsius) |
The various flags are introduced into the dataset to provide additional information about certain occasions when bar trading was especially busy. The ten records with the largest beverage sales all have various combinations of these flags.
We are looking to build a regression model that will quantify the extent that the predictor variables are influencing the response variable. In this case the response variable is bev\\\_quant. The predictor variables of interest are the continuous variables bpm, n\\\_events, precip, relhum, temp and the categoricals is\\\_pre\\\_xmas, is\\\_mon\\\_pubhol, is\\\_pubhol, is\\\_pubhol\\\_eve, and is\\\_happy\\\_hr. This relationship is going to involve some assumptions about the randomness inherent in any set of observations.
The Generalized Linear Model
For count data the appropriate model is a discrete probability distribution over the whole numbers 0, 1, 2, …, of which the simplest distribution is the Poisson. Thus we consider the number of drinks purchased per hour to be a random variable from a Poisson distribution whose mean rate is potentially influenced by the factors above. The level of detail in the model could be increased if we knew the number of hotel patrons present throughout the hour, but unfortunately that data is not available.
When modelling count data, linear regression (Ordinary Least Squares) is not appropriate. It requires the response variable to be a continuous quantity without restrictions on its range, whereas count data must have non-negative integer values.
For such data Generalized Linear Models (GLM) provide a better solution. A GLM may be described in the following way.
- The data is represented as $N$ observations $y_i$ of the response variable, each with $p-1$ associated values $x_{ij}$ of the predictor variables;
- The linear part of the model is the vector product $x^T_i \beta$, where $x_i$ is the augmented $p \times 1$ vector of the $x_{ij}$ and $\beta$ is the $p \times 1$ vector of unknown model coefficients;
- The model uses a distribution of the exponential dispersion models, parameterized by its mean $\mu$, to describe the response variable. Examples of such distributions are the Normal, Binomial, Poisson and Negative Binomial distributions. For each distinct combination of values of the predictors we allow a different mean $\mu_i$ and assume the associated $y_i$ are sampled from that distribution.
- Each $\mu_i$ is related to the linear combination of predictor variables through a link function $g(\mu)$ as expressed in the equation
The GLM regression finds the most likely value of $\beta$ for the given data. In most cases this requires an iterative algorithm that maximises the log-likelihood function.
Note that OLS is a particular case of GLM in which the Normal distribution is assumed for $y_i$ and the identity link function $g(\mu_i) = \mu_i$ is used.
Poisson regression
When applied to a Poisson response variable, the GLM is called Poisson regression. The usual link function in this case is the natural logarithm function, although other choices are possible provided the linear function $x^T_i \beta$ does not map the data beyond the domain of $g^{-1}$.
The variance of a Poisson random variable is equal to the mean, so we expect this to be true for our data if the underlying distribution truly is Poisson. Cases where the variance exceeds the mean, referred to as overdispersion, can arise when one or more unobserved variables are contributing to the mean but are, for whatever reason, not included in the model. (If those variables were to be included in the regression, the variance would be reduced as the contribution to the residuals caused by their absence would be eliminated.) More rarely, the variance may be less than the mean, in which case the data is said to be underdispersed. Over- and underdispersion are both indications that the Poisson model is inappropriate, as the standard errors are under- or over-estimated, respectively, and an alternate model should be sought. Statsmodels provides information about the goodness of fit that may be used to detect these cases.
We begin by applying Poisson regression as shown in the script below. The variable data is the DataFrame with the selected data. The patsy formula notation simplifies construction of the design matrices required by Statsmodels. This includes creating a column of 1’s in the predictor matrix, predictors, to allow for an intercept parameter in the model. By wrapping the names of the flag columns in “C(…)” we are indicating they are categoricals.
Poisson regression
| from patsy import dmatrices | |
| import statsmodels.api as sm | |
| formula = “””bev_quant ~ bpm + n_events + precip + relhum + temp + C(is_happy_hr) | |
| + C(is_mon_pubhol) + C(is_pre_xmas) + C(is_pubhol) + C(is_pubhol_eve)””” | |
| response, predictors = dmatrices(formula, data, return_type=’dataframe’) | |
| po_results = sm.GLM(response, predictors, family=sm.families.Poisson()).fit() | |
| print(po_results.summary()) |
view rawpoisson-regr.py hosted with ❤ by GitHub
| from patsy import dmatrices | |
| import statsmodels.api as sm | |
| formula = “””bev_quant ~ bpm + n_events + precip + relhum + temp + C(is_happy_hr) | |
| + C(is_mon_pubhol) + C(is_pre_xmas) + C(is_pubhol) + C(is_pubhol_eve)””” | |
| response, predictors = dmatrices(formula, data, return_type=’dataframe’) | |
| po_results = sm.GLM(response, predictors, family=sm.families.Poisson()).fit() | |
| print(po_results.summary()) |
view rawpoisson-regr.py hosted with ❤ by GitHub
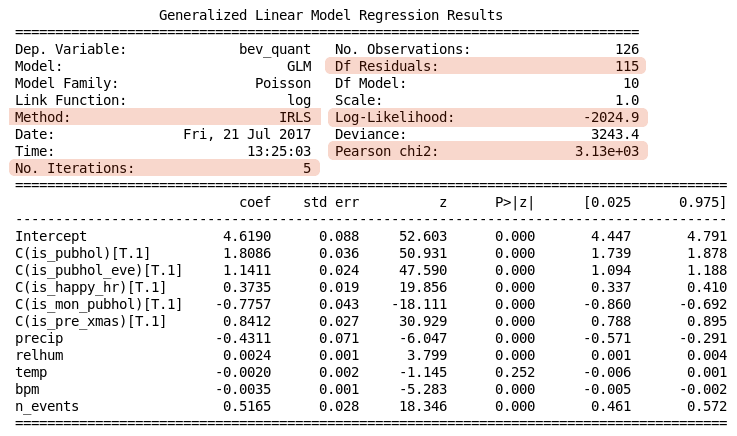
The output from this script is shown in Table 2 below. Notice there is an upper table with general details about the regression calculation, and a lower table with the calculated regression coefficients and some statistics about them. Initially we are interested in the upper table, specifically in those fields that are highlighted.
Table 2. Poisson regression summary
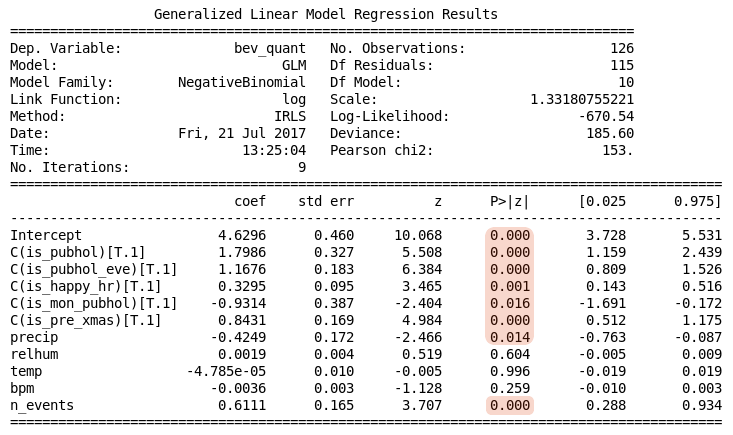
The output indicates there were 5 iterations of the IRLS algorithm before it converged on a solution with the log-likelihood of -2025. The particular value of the log-likelihood is not important, rather we should be concerned about how this changes with adjustments to the model. Together this information suggests there was no difficulty finding a solution.
The adequacy of a GLM is generally determined from its residuals, which are scaled measures, $r_i$, of the amounts by which the response observations differ from their respective means:
where $s_i$ represents a form of standard error depending on the particular kind of residual (Pearson, Deviance, etc). For our purposes the Pearson residuals are sufficient. The sum of the squares of these residuals can be used as an aggregate statistic to assess the adequacy of the model. For the Pearson statistic Statsmodels reports this as Pearson chi2.
To better understand the data and the adequacy of the Poisson model, we plot the Pearson residuals against their associated fitted means $\mu_i$ in Figure 2A. This shows about 75% of the data falls into one of two vertical clusters with means in the ranges [60, 89] or [90, 119]. The remaining data are spread relatively sparsely across the range of means. Note that this compression of data towards the lower end of the scale is partly due to the log link function. The Pearson residual is scaled to compensate for the non-uniform variance, so we would expect a more uniform vertical scattering than appears here, which may be an indicator of overdispersion. The greatest divergence from the model appears to be in the mid-range [150, 350]. Figure 2B is discussed below.
Figure 2. Poisson residuals and dispersion

The Pearson statistic provides an estimate of the data’s dispersion. When the data is drawn from a Poisson distribution with sufficient samples the ratio Pearson chi2 / Df Residuals is approximately 1; for observed data a ratio less than 1 implies underdispersion and more than 1 implies overdispersion. Data that is underdispersed requires a zero-inflated model, which is not directly implemented by Statsmodels. In this case the result, 27.2, suggests overdispersion, which is amenable to Negative Binomial regression. We explore that below using Statsmodels.
The upper results table shown in Table 2 reports other details which may also be used to assess the adequacy of the model in various situations. The deviance is calculated from the log-likelihood and may be used in various statistical tests.
The lower results table shown in Table 2 reports details of the model parameters, assuming that the model is appropriate. Given the overdispersion finding in this case, these details should be considered unreliable.
Negative Binomial regression
The Negative Binomial distribution is a generalization of the Poisson distribution. Although typically introduced in terms of a sequence of Bernoulli trials, an alternative formulation allows it to be considered as a mixture distribution in which samples are drawn from a Poisson distribution whose mean is itself a random variable drawn from a Gamma distribution. This allows the Negative Binomial distribution to use a continuous positive dispersion parameter, $\alpha$ to specify the extent by which the distribution’s variance exceeds its mean. Specifically, the variance can be described by the equation
from which we see that as $\alpha$ tends to zero, the variance approaches the mean, which is the behaviour of the Poisson distribution.
The $\alpha$ value must be specified as an input to the model, raising a question about how to select the right value. There is little guidance on this – the Statsmodels documentation suggests a value in the range [.01, 2] is permissible, without elaborating. However Cameron and Trivedi describe a test for under- or overdispersion of a Poisson model which estimates $\alpha$ and, for small datasets, can be a more reliable indicator than the Pearson statistic used above. Unfortunately this test is not part of Statsmodels, but is easily implemented. To perform the test we take the fitted means $\mu_i$ from a Poisson model and perform an OLS regression (without an intercept) for the auxiliary model
where the left-hand side is treated as the response variable, $\alpha$ is the unknown model coefficient and $\epsilon_i$ is an error term representing random variation in the responses. The fitted coefficient $\alpha$ has a $t$ distribution, from which we can construct a confidence interval that indicates the extent of dispersion and provides a suitable parameter value for the Negative Binomial regression. The script below performs this calculation for a 95% confidence interval using Statsmodels’ OLS feature and the results from the previous Poisson regression. Note the “- 1” term in the regression formula which instructs patsy to remove the column of 1’s from the design matrix.
Dispersion calculation for Poisson regression
| import statsmodels.formula.api as smf | |
| def ct_response(row): | |
| “Calculate response observation for Cameron-Trivedi dispersion test” | |
| y = row[‘bev_quant’] | |
| m = row[‘bev_mu’] | |
| return ((y – m)**2 – y) / m | |
| ct_data = data.copy() | |
| ct_data[‘bev_mu’] = po_results.mu | |
| ct_data[‘ct_resp’] = ct_data.apply(ct_response, axis=1) | |
| # Linear regression of auxiliary formula | |
| ct_results = smf.ols(‘ct_resp ~ bev_mu – 1’, ct_data).fit() | |
| # Construct confidence interval for alpha, the coefficient of bev_mu | |
| alpha_ci95 = ct_results.conf_int(0.05).loc[‘bev_mu’] | |
| print(‘\nC-T dispersion test: alpha = {:5.3f}, 95% CI = ({:5.3f}, {:5.3f})’ | |
| .format(ct_results.params[0], alpha_ci95.loc[0], alpha_ci95.loc[1])) |
view rawpoisson-dispersion.py hosted with ❤ by GitHub
| import statsmodels.formula.api as smf | |
| def ct_response(row): | |
| “Calculate response observation for Cameron-Trivedi dispersion test” | |
| y = row[‘bev_quant’] | |
| m = row[‘bev_mu’] | |
| return ((y – m)**2 – y) / m | |
| ct_data = data.copy() | |
| ct_data[‘bev_mu’] = po_results.mu | |
| ct_data[‘ct_resp’] = ct_data.apply(ct_response, axis=1) | |
| # Linear regression of auxiliary formula | |
| ct_results = smf.ols(‘ct_resp ~ bev_mu – 1’, ct_data).fit() | |
| # Construct confidence interval for alpha, the coefficient of bev_mu | |
| alpha_ci95 = ct_results.conf_int(0.05).loc[‘bev_mu’] | |
| print(‘\nC-T dispersion test: alpha = {:5.3f}, 95% CI = ({:5.3f}, {:5.3f})’ | |
| .format(ct_results.params[0], alpha_ci95.loc[0], alpha_ci95.loc[1])) |
view rawpoisson-dispersion.py hosted with ❤ by GitHub
This script prints the following result, confirming the earlier assessment of overdispersion (0 lies outside the confidence interval) and providing an estimate for $\alpha$.
C-T dispersion test: alpha = 0.150, 95% CI = (0.103, 0.198)
Figure 2B shows the C-T response values plotted against the fitted means, together with the fitted line. Once again this shows the influence of the outlying data points on the dispersion estimate.
The script below performs Negative Binomial regression on the same dataset as before. Its output is shown in Table 3 below.
Negative Binomial regression
| f = “””bev_quant ~ precip + relhum + temp + bpm + n_events + C(is_pubhol) | |
| + C(is_pubhol_eve) + C(is_happy_hr) + C(is_mon_pubhol) + C(is_pre_xmas)””” | |
| response, predictors = dmatrices(f, data, return_type=’dataframe’) | |
| nb_results = sm.GLM(response, predictors, | |
| family=sm.families.NegativeBinomial(alpha=0.15)).fit() | |
| print(nb_results.summary()) |
view rawnbinom-regr.py hosted with ❤ by GitHub
| f = “””bev_quant ~ precip + relhum + temp + bpm + n_events + C(is_pubhol) | |
| + C(is_pubhol_eve) + C(is_happy_hr) + C(is_mon_pubhol) + C(is_pre_xmas)””” | |
| response, predictors = dmatrices(f, data, return_type=’dataframe’) | |
| nb_results = sm.GLM(response, predictors, | |
| family=sm.families.NegativeBinomial(alpha=0.15)).fit() | |
| print(nb_results.summary()) |
view rawnbinom-regr.py hosted with ❤ by GitHub
Table 3. Negative Binomial regression summary
The information in the upper table indicates a better fit has been achieved, as would be expected. A larger value for the log-likelihood has been achieved and the ratio Pearson chi2 / Df Residuals is now slightly greater than 1.
The lower table provides the detail of the regression coefficients, including p-values for testing whether the coefficients are different from zero as well as 95% confidence intervals. Statistical significance at the 5% level is indicated (P>|z| < 0.05) for is\\\_pubhol, is\\\_pubhol\\\_eve, is\\\_happy\\\_hr, is\\\_mon\\\_pubhol, is\\\_pre\\\_xmas, precip, and n\\\_events, with is\\\_mon\\\_pubhol and precip having a negative contribution. The coefficients for the other variables are not significantly different from zero.
Advantages of Statsmodels
In this article I have shown how GLM regression models can be implemented in just a few lines of Python code using Statsmodels. What may not be apparent here is that in addition to being concise, the Statsmodels API is also quite flexible. It has a modular, object-oriented design that allows a broad range of specific model variants to be defined by composing a model with a probability distribution and a link function. Statsmodels also uses Python’s convenient syntax for default function arguments and includes sensible defaults that address typical usage. Many functions have default arguments that give the user fine control over the algorithm details if desired. As shown here the regression results can be viewed as a tabular summary or individual details can be accessed as attributes or methods of the results object.
One alternative to using Statsmodels for some GLM problems would be to use Scikit-learn, which offers OLS (and regularized variants) and logistic regression, but does not support count data with the discrete distributions offered by Statsmodels. Scikit-learn focuses on classification and regression models for high-dimensional data (i.e. having many predictors), offering various forms of regularization and algorithms suitable for very large datasets, but not the statistical features offered by Statsmodels.
Conclusion
As indicated earlier, the number of hotel patrons was not available but if it were available this would probably have changed the analysis and results. Most of the categorical predictors have at least part of their effect by increasing the patronage. The effect of this cuts across the predictors and thus requires a different representation, as a $N \times 1$ vector of “exposure” variables, which Statsmodels can accommodate.
This exploration has demonstrated both the ease and capability of the Statsmodels GLM module. For a user having some familiarity with OLS regression and once the data is in a pandas DataFrame, powerful regression models can be constructed in just a few lines of code. Attention must be paid to the results to determine whether the model is appropriate for the data, but Statsmodels provides sufficient information to make that judgement.
References
- Seabold, Skipper, and Josef Perktold. “Statsmodels: Econometric and statistical modeling with python.” Proceedings of the 9th Python in Science Conference. 2010.
- Dobson, A. J. and Barnett, A. G. “An Introduction to Generalized Linear Models.” Third Edition, Chapman and Hall/CRC. 2008.
- Cameron, A. C. and Trivedi, P. K. “Essentials of Count Data Regression,” (2009). Retrieved from https://cameron.econ.ucdavis.edu/research/CTE01preprint.pdf

